A Review of Current Cell Annotation Systems for Histopathology Images
DOI:
https://doi.org/10.37934/ard.135.1.88100Keywords:
Cell annotation, cell labelling, cell segmentation, nuclei segmentation, system, histopathology, digital pathologyAbstract
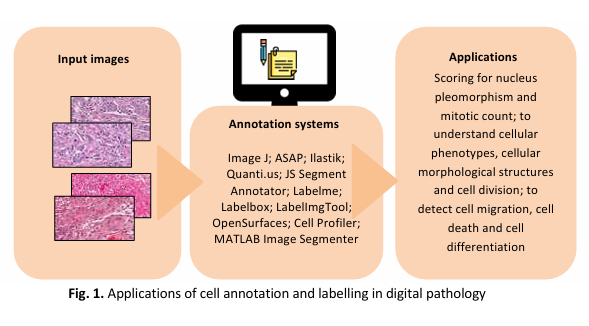
Histopathology techniques offer a unique way to study the structural and functional characteristics of biological model systems such as cultured cells, tissues and organoids. As the field of histopathology advances and more complex properties of living organisms are revealed through novel assays, there is a growing need for image analysis methods that are robust and easy to use. In many histopathology image analysis workflows, the first step is to segment cell nuclei, as they serve as the fundamental block for identifying individual cells in histopathology images. Such methods are crucial in a range of research studies, from counting cells and tracking moving populations to localizing proteins, classifying phenotypes and profiling treatments. Here, this review intended to provide an updated cell annotation system for histopathology images using the predetermined search strings and a set of inclusion criteria. Accordingly, 11 cell annotation systems (i.e., Image J, ASAP, Ilastik, Quanti.us, JS Segment Annotator, Labelme annotation tool, Labelbox, LabelImgTool, OpenSurfaces Segmentation UI, Cell Profiler and MATLAB Image Segmenter) was included. An in-depth discussion on the background of each included system was provided in this review alongside a brief comparison across different cell annotation systems.
Downloads